Original-Text
Thurstone
"trapezoid
population
In previous studies of factorial theory it has been found
useful to illustrate the principles by means of a population of simple
physical objects or geometrical figures. The box population was used to
illustrate three correlated factors and their physical interpretation.
In the present case we want four factors in the first-order domain, which,
by their correlations of unit rank, determine a general second-order factor.
The correlations of three variables can nearly always be accounted for
by a single factor, and hence it seems better to choose a four-dimensional
system in which the existence of a second-order general factor is more
clearly indicated by the unit rank of the correlations of four primary
factors. For the present physical illustration we have chosen a population
of trapezoids whose shapes are determined by four primary parameters or
factors.
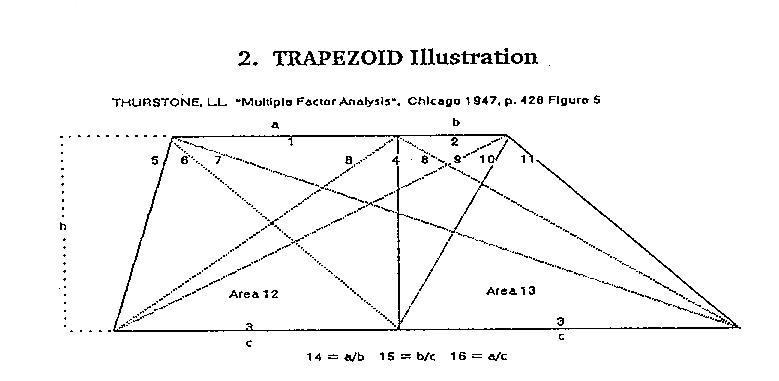
The measurements on the trapezoids are indicated in Figure
5. The base line is bisected, and the length of esch half is denoted by
the parameter c. An ordinate is erected at this mid-point, and its length
is h. This ordinate divides the top section into two parts, which are denoted
a and b as shown. These four parameters, a, b, c, and h, completely determine
the figure. The test battery was represented by sixteen measurements, wEich
are drawn in the figure. The parameters a, b, c, and h are given code numbers
1, 2, 3, and 4, respectively. Variables (12) and (13) are the two areas
as shown. The sum of (12) and
(13) equals the total area of the trapezoid. In general, each of these
measurements is a function of two or three of the parameters but not of
all four of them, and hence we should expect a simple structure in this
[p. 428] set of measurements. There is a rather general impression that
a simple structure is necessarily confined to the positive manifold. In
order to offset this impression we included here three additional measures,
which extend the simple structure beyond the positive manifold. These three
additional measures are as follows:
14 = (1)/(2) = a/b, 15 = (2)/(3) = b/c, 16 = (1)/(3) = a/c .
These three measures will necessarily introduce negative
saturations on some of the basic factors.
In Table 6 we have a list of dimensions for a set of thirty-two
trapezoids. These will constitute the trapezoid population. Each figure
was drawn to
FIGURE 5
scale on cross-section paper, and then the sixteen measurements
were made on each figure. These constituted the test scores for the present
example. In setting up the dimensions of Table 6 the numbers were not distributed
entirely at random. To do so would tend to make the correlations between
the four basic parameters, a, b, c, and h, approach zero, and this would
lead to an orthogonal simple structure in which there would be no provocation
to investigate a second-order domain. The manner in which the generating
conditions of the objects determine the factorial results will be discussed
in a later section. Table 6 was so constructed that, in addition to the
four basic parameters, there was also a size factor, which functioned as
a second-order parameter in determining correlation between the four primary
factors in generating the figures.
The product-moment correlations between the sixteen measurements
for the thirty-two objects were computed, and these are listed in Table
7. This [p. 429] correlation matrix was factored by the group centroid
method, and the resulting factor matrix F is shown in Table 8. The fourth-factor
residuals are listed in Table 9, which indicates that the residuals are
vanishingly small." |
3.
Übersicht der Ergebnisse und bibliographische Hinweise
THURSTONE, L. L. (USA: University Of Chicago) "Multiple
Factor Analysis" Chicago 1947, p.427-436 "A trapezoid
population", p. 431 Table 7 'Correlation Matrix' a n d THURSTONE,
L. L. "SECOND-ORDER FACTORS" Psychometrika 9.2, 1944, p.96
Table 7 Correlation Matrix.
Bemerkung-1: Die Matrix enthält zwei Druckfehler: r14,16 und r15,16
sollten positiv sein. Man kann hier studieren, wie sich ein Druckfehler
hinsichtlich des Vorzeichens zweier Korrelationskoeffizienten auf die numerische
Stabilität auswirkt.
(6=12f) THPMF16.K16 raw scores measured with 2-digit input
accuray read
Samp _Ord_
MD_
NumS_
Condition_
Determinant_HaInRatioR_
OutInK_
Norm
_
C_Norm
32 16 0 --4
3711 -3.67 D-20 3.12 D-23
347573 0(8) -1(-1)
Zum Widerspruch negative Determinante und geradzahlig negative Eigenwerte,
die zu einer positiven Determinante führen müßten, siehe
bitte unten.
Dieselbe wie obige Matrix ohne Druckfehler und aus Thurstone's Buch
(1=12a) TH43116.K16 raw scores measured with 2-digit input
accuray read
Samp _Ord_
MD_
NumS_
Condition_
Determinant_HaInRatioR_
OutInK_
Norm
_
C_Norm
32 16 0 --4
2992 6.36 D-21 1.43 D-13
79208 2D-3(9) -1(-1)
Korrigierte Version: TRAPEZ32.D16
with 17-digit-input accuracy calculed
Samp _Ord_
MD_NumS_Condition_Determinant_HaInRatioR_
OutInK_
NormC_
Norm
32 16 0 -
502896.5 0
3.81D-31 17952 0(8) 0.006(7)
TRAPEZ32.D16 korrigiert with 4-digit-input
accuracy read
Samp _Ord_
MD_
NumS_
Condition_
Determinant_HaInRatioR_
OutInK_
Norm _
C_Norm
32 16 0 --1
594867.1 0
2.25D-32 17952 0(8) -1(-1)
TRAPEZ32.D16 korrigiert with 3-digit-input
accuracy read
Samp _Ord_
MD_
NumS_
Condition_
Determinant_HaInRatioR_
OutInK_
Norm
_
C_Norm
32 16 0 --2
2.1D+6 0
2.81D-43 6698.1 0(8) -1(-1)
In der korrigierten Version ist die Konditionszahl rund 8x größer
und ein negativer Eigenwert kommt hinzu. Dies illustriert ganz gut, wie
numerische Instabilität wirkt.
TRAPEZ32.D16 korrigiert with 2-digit-input
accuracy read
Samp _Ord_
MD_
NumS_
Condition_
Determinant_HaInRatioR_
OutInK_
Norm
_
C_Norm
32 16 0 --4
14892.5 0
7.01D-18 17952 0(8)
-1(-1)
4. Detail Analysen
(1)
Values measured by Thurstone with 2-digit input accuray read
THURSTONE, L. L. (USA: University Of Chicago) "Multiple
Factor Analysis" Chicago 1947, p.427-436 "A trapezoid
population", p. 431 Table 7 'Correlation Matrix'
Samp Or MD NumS Condit Determinant
HaInRatio R_OutIn K_Norm C_Norm
32 16 0 --4
2992 6.36 D-21 1.43 D-13 79208
2D-3(9) -1(-1)
********** Summary of standard correlation
matrix analysis ***********File = TH431_16.K16 N-order=
16 N-sample= 32 Rank= 16 Missing data = 0
Positiv Definit=Cholesky successful________= No with
4 negative eigenvalue/s
HEVA: Highest eigenvalue abs.value_________= 10.288
LEVA: Lowest eigenvalue absolute value_____= 0.003438778
CON: Condition number HEVA/LEVA___________~= 2991.69
DET: Determinant original matrix___________= D-21 6.359
HAC: HADAMARD condition number_____________= D-27 1.481
HCN: Heuristic condition |DET|CON__________= D-24 2.125
D_I: Determinant Inverse absolute value____= 1.5724911329896272D+20
HDA: HADAMARD Inequation absolute value___<= 1.0988050960598554D+33
HIR: HADAMARD RATIO: D_I / HDA ____________= 1.4310919549138755D-13
Highest inverse positive diagonal value____= 60.001125634
thus multiple r( 7.rest)_________________= .99163181
and 1 multiple r > .99
Highest inverse negative diagonal value____= -1.762489944
thus multiple r( 12.rest)_________________= 1.251950128
(!)
and there are 5 multiple r > 1 (!)
Maximum range (upp-low) multip-r( 1.rest)_= .257
LES: Numerical stability analysis:
Maximum range input x(upper)-x(lower)____=
0.009
Maximum range output x(upper)-x(lower)____= 712.87197
Ratio maximum range output / input _______= D +4
7.9
Mean absolute value of ranges output _____= 335.51821
Ratio mean range output/ mean range input_= D +4
3.7
Sigma of mean (abs. value range output)___= 244.66269
Ncor L1-Norm L2-Norm Max
Min m|c| s|c|
Ncomp M-S S-S
256 156 10.79
1 -.84 .61
.527 120 .218 .244
class boundaries and distribution of the correlation-coefficients
-1 -.8 -.6 -.4 -.2 0
.2 .4 .6 .8 1
2 8 12
20 12 18 24 32
46 82
Original input data with 2-digit-accuracy and read with 2-digit-accuracy
(for control here the analysed original matrix):
1 .5 .5 .32
.29 .58 .72 .49 .58 .45 .31 .66
.53 .76 -.35 .11
.5 1 .5 .32
.36 .42 .57 .49 .74 .67 .33 .54
.64 -.16 -.14 -.23
.5 .5 1 .32
.52 .42 .88 .82 .9 .45 .3
.78 .75 .19 -.84 -.72
.32 .32 .32 1 .95
.96 .65 .8 .61 .91 .98 .78
.82 .12 -.22 -.15
.29 .36 .52 .95 1
.9 .75 .9 .75 .9 .94
.84 .89 .05 -.37 -.31
.58 .42 .42 .96 .9 1
.78 .83 .7 .92 .94 .86 .86
.34 -.29 -.09
.72 .57 .88 .65 .75 .78
1 .95 .95 .74 .64 .95 .91
.39 -.69 -.46
.49 .49 .82 .8 .9
.83 .95 1 .93 .83 .78 .94
.95 .19 -.64 -.52
.58 .74 .9 .61 .75 .7
.95 .93 1 .79 .6 .9
.93 .11 -.64 -.57
.45 .67 .45 .91 .9 .92
.74 .83 .79 1 .9 .83
.9 .01 -.22 -.21
.31 .33 .3 .98 .94 .94
.64 .78 .6 .9 1 .77
.8 .11 -.12 -.09
.66 .54 .78 .78 .84 .86
.95 .94 .9 .83 .77 1
.97 .34 -.59 -.39
.53 .64 .75 .82 .89 .86
.91 .95 .93 .9 .8 .97 1
.12 -.52 -.44
.76 -.16 .19 .12 .05 .34 .39
.19 .11 .01 .11 .34 .12 1
-.28 .34
-.35 -.14 -.84 -.22 -.37 -.29 -.69 -.64 -.64 -.22 -.12 -.59 -.52
-.28 1 .76
.11 -.23 -.72 -.15 -.31 -.09 -.46 -.52 -.57 -.21 -.09 -.39
-.44 .34 .76 1
i.Eigenvalue Cholesky i.Eigenvalue
Cholesky i.Eigenvalue Cholesky
1. 10.28775 1
2. 2.36742 .866
3. 1.89721 .8165
4. 1.12211 .92
5. .16865 .1697
6. .07716 -5.6D-3
7. .06717 -.5981
8. .01777 -1.5864 9.
.01327 -2.2646
10. 9.35D-3 -2.7013 11. 4.55D-3
-3.0387 12. 3.44D-3 -4.5821
13.-3.69D-3 -5.6998 14.-7.39D-3
-.4257 15.-.01038 -1.9947
16.-.01438 -1.8594
The matrix is not positive definit. Cholesky decomposition
is not successful (for detailed information Cholesky's diagonalvalues are
presented).
(2)
Values are computed and not measured with 17-digit-input-accuracy
& calculed (k)
TRAPEZ32.D16 korrigiert with 17-digit-input
accuracy read
Samp Or MD NumS Condit Determinant
HaInRatio R_OutIn K_Norm C_Norm
32 16 0 -
502896.5 0
3.81D-31 17952 0(8)
0.006(7)
********** Summary of standard correlation matrix
analysis ***********
File = TRAPEZ32.D16 N-order= 16 N-sample= 32
Rank= 16 Missing data = 0
Positiv Definit=Cholesky successful________= Yes with 0 negat.
eigenvalue/s
HEVA: Highest eigenvalue abs.value_________=
10.218631637726756
LEVA: Lowest eigenvalue absolute value_____=
2.0319552205460591D-5
CON: Condition number HEVA/LEVA___________~=
502896.49763938421
DET: Determinant original matrix (OMIKRON)_=
1.645078683984506D-29
DET: Determinant (CHOLESKY-Diagonal^2)_____=
1.6450786839845063D-29
DET: Determinant (PESO-CHOLESKY)___________=
1.6450786839845063D-29
DET: Determinant (product eigenvalues)_____=
1.6450786839814124D-29
DET: Determ.abs.val.(PESO prod.red.norms)__=
1.6450786839844999D-29
HAC: HADAMARD condition number_____________=
4.2714963680318048D-36
HCN: Heuristic condition |DET|CON__________=
3.271207279642172D-35
D_I: Determinant Inverse absolute value____=
6.0787365962211835D+28
HDA: HADAMARD Inequality absolute value___<=
1.5933927985332463D+59
HIR: HADAMARD RATIO: D_I / HDA ____________=
3.8149642710929763D-31
Highest inverse positive diagonal value____=
25369.293808947
thus multiple r( 9.rest)_________________=
.999980291
and 15 multiple r > .99
There are no negative inverse diagonal values.
Maximum range (upp-low) multip-r( 2.rest)_=
.171
LES: Numerical stability analysis:
Ratio maximum range output / input _______=
17951.973072121561
PESO-Analysis correlation least Ratio RN/ON=
1D-5 (<-> Angle = 0 )
Number of Ratios correlation RN/ON < .01__ =
8
PESO-Analysis Cholesky least Ratio RN/ON__ =
6.278D-3 (<-> Angle = .36 )
Number of Ratios Cholesky RN/ON < .1 _____ =
7
Ncor L1-Norm L2-Norm Max
Min m|c| M|c| N_comp
s-S S-S
256 154.7 10.72
1 -.83 .578
.28 7140 .323 .231
class boundaries and distribution of the correlation coefficients
-1 -.8 -.6 -.4 -.2 0
.2 .4 .6 .8 1
2 10 10
20 14 16 24 32
46 82
i.Eigenvalue Cholesky i.Eigenvalue
Cholesky i.Eigenvalue Cholesky
1. 10.21863 1
2. 2.3971 .866
3. 1.84973 .8165
4. 1.15395 .922
5. .19044 .164
6. .08281 .0535
7. .06998 .0597
8. .01708 .0158
9. 8.13D-3 .0257
10. 6.88D-3 .0343 11.
2.47D-3 .0976 12. 2.13D-3
.1371
13. 4.5D-4 .0501
14. 1.6D-4 .1921
15. 4D-5 .0722
16. 2D-5 .0915
Cholesky decomposition successful, thus the matrix is (semi)
positive definit.
Eigenvalues in per cent of trace = 16
1 .6387 2 .1498 3 .1156
4 .0721 5 .0119 6 5.2D-3
7 4.4D-3 8 1.1D-3 9 5D-4 10
4D-4 11 2D-4 12 1D-4
13 0 14 0
15 0 16 0
[Intern: analysed: 10/22/02 22:51:56 PRG version
05/24/94 MA9.BAS
File = C:\OMI\NUMERIK\MATRIX\SMA\TRAPEZ32\TRAPEZ32.SMA
with data from C:\OMI\NUMERIK\MATRIX\SMA\TRAPEZ32\TRAPEZ32.D16
Date: 10/22/02 Time:22:51:56]
(3)
Values are computed and not measured with 4-digit-input-accuracy read (k)
TRAPEZ32.D16 korrigiert with 4-digit-input
accuracy read
Samp _Ord_
MD_
NumS_
Condition_
Determinant_HaInRatioR_
OutInK_
Norm _
C_Norm
32 16 0 --1
594867.1 0
2.25D-32 17952 0(8)
-1(-1)
********** Summary of standard correlation matrix
analysis ***********
File = TRAPEZ32.D16 N-order= 16 N-sample= 32
Rank= 16 Missing data = 0
Positiv Definit=Cholesky successful________= No with 1 negat.
eigenvalue/s
HEVA: Highest eigenvalue abs.value_________=
10.218634101122853
LEVA: Lowest eigenvalue absolute value_____=
1.7178012311068469D-5
CON: Condition number HEVA/LEVA___________~=
594867.0845077102
DET: Determinant original matrix (OMIKRON)_= -2.4694269803906613D-29
DET: Determinant (CHOLESKY-Diagonal^2)_____= -999 (not
positive definit)
DET: Determinant (PESO-CHOLESKY)___________= -999 (not
positive definit)
DET: Determinant (product eigenvalues)_____= -2.4694269803881388D-29
DET: Determ.abs.val.(PESO prod.red.norms)__=
2.4694269803906669D-29
HAC: HADAMARD condition number_____________=
6.4120207821971545D-36
HCN: Heuristic condition |DET|CON__________=
4.1512247772698113D-35
D_I: Determinant Inverse absolute value____=
4.0495224517300804D+28
HDA: HADAMARD Inequality absolute value___<=
1.7987156414681396D+60
HIR: HADAMARD RATIO: D_I / HDA ____________=
2.2513411004892343D-32
Highest inverse positive diagonal value____=
25208.251484587
thus multiple r( 9.rest)_________________=
.999980165
and 12 multiple r > .99
Highest inverse negative diagonal value____= -153.889894333
thus multiple r( 10.rest)_________________=
1.003243815 (!)
and there are 2 multiple r > 1 (!)
Maximum range (upp-low) multip-r( 2.rest)_=
.171
LES: Numerical stability analysis:
Ratio maximum range output / input _______=
17951.973072121561
PESO-Analysis correlation least Ratio RN/ON=
8D-6 (<-> Angle = 0 )
Number of Ratios correlation RN/ON < .01__ =
8
PESO-Analysis Cholesky least Ratio RN/ON__ = (Not positiv definit)
Ncor L1-Norm L2-Norm Max
Min m|c| M|c| N_comp
s-S S-S
256 154.7 10.72
1 -.83 .578
.28 7140 .323 .231
class boundaries and distribution of the correlation coefficients
-1 -.8 -.6 -.4 -.2 0
.2 .4 .6 .8 1
2 10 10
20 14 16 24 32
46 82
i.Eigenvalue Cholesky i.Eigenvalue
Cholesky i.Eigenvalue Cholesky
1. 10.21863 1
2. 2.39706 .866
3. 1.84972 .8165
4. 1.15396 .922 5.
.19044 .1638 6.
.08281 .054
7. .07002 .0611
8. .01706 .0165
9. 8.2D-3 .025
10. 6.84D-3 .029
11. 2.44D-3 -1.4D-3 12. 2.22D-3 -.4691
13. 4.1D-4 -1.5393 14. 2.4D-4
-.0422 15. 2D-5 -.7613
16.-5D-5 -1.0652
The matrix is not positive definit. Cholesky decomposition
is not success-
Eigenvalues in per cent of trace = 16
1 .6387 2 .1498 3 .1156
4 .0721 5 .0119 6 5.2D-3
7 4.4D-3 8 1.1D-3 9 5D-4 10
4D-4 11 2D-4 12 1D-4
13 0 14 0
15 0 16 0
[Intern: analysed: 10/22/02 23:36:22 PRG version
05/24/94 MA9.BAS
File = C:\OMI\NUMERIK\MATRIX\SMA\TRAPEZ32\TRAPEZ32.SMA
with data from C:\OMI\NUMERIK\MATRIX\SMA\TRAPEZ32\TRAPEZ32.D16
Date: 10/22/02 Time:23:36:22]
(4)
Values are computed and not measured with 3-digit-input-accuracy read (k)
TRAPEZ32.D16 korrigiert with 3-digit-input
accuracy read
Samp Or MD NumS Condit Determinant
HaInRatio R_OutIn K_Norm C_Norm
32 16 0 --2 2.1D+6
0 2.81D-43
6698.1 0(8) -1(-1)
********** Summary of standard correlation matrix
analysis ***********
File = TRAPEZ32.D16 N-order= 16 N-sample= 32
Rank= 16 Missing data = 0
Positiv Definit=Cholesky successful________= No with 2 negat.
eigenvalue/s
HEVA: Highest eigenvalue abs.value_________=
10.217959342460799
LEVA: Lowest eigenvalue absolute value_____=
4.6539581285637635D-6
CON: Condition number HEVA/LEVA___________~=
2195541.7432202202
DET: Determinant original matrix (OMIKRON)_=
3.4172590164546888D-28
DET: Determinant (CHOLESKY-Diagonal^2)_____= -999 (not
positive definit)
DET: Determinant (PESO-CHOLESKY)___________= -999 (not
positive definit)
DET: Determinant (product eigenvalues)_____=
3.4172590164729643D-28
DET: Determ.abs.val.(PESO prod.red.norms)__=
3.4172590164547137D-28
HAC: HADAMARD condition number_____________=
8.8832239172445477D-35
HCN: Heuristic condition |DET|CON__________=
1.5564536757304214D-34
D_I: Determinant Inverse absolute value____=
2.9263219299000407D+27
HDA: HADAMARD Inequality absolute value___<=
1.0390704560685637D+70
HIR: HADAMARD RATIO: D_I / HDA ____________=
2.8162882630424298D-43
Highest inverse positive diagonal value____=
28.70356303
thus multiple r( 14.rest)_________________=
.98242614
Highest inverse negative diagonal value____= -22.99665269
thus multiple r( 11.rest)_________________=
1.021510935 (!)
and there are 15 multiple r > 1 (!)
Maximum range (upp-low) multip-r( 2.rest)_=
.101
LES: Numerical stability analysis:
Ratio maximum range output / input _______=
6698.1075669521929
PESO-Analysis correlation least Ratio RN/ON=
3D-6 (<-> Angle = 0 )
Number of Ratios correlation RN/ON < .01__ =
8
PESO-Analysis Cholesky least Ratio RN/ON__ = (Not positiv definit)
Ncor L1-Norm L2-Norm Max
Min m|c| M|c| N_comp
s-S S-S
256 154.7 10.72
1 -.83 .578
.28 7140 .323 .231
class boundaries and distribution of the correlation coefficients
-1 -.8 -.6 -.4 -.2 0
.2 .4 .6 .8 1
2 10 10
20 14 16 24 32
46 82
i.Eigenvalue Cholesky i.Eigenvalue
Cholesky i.Eigenvalue Cholesky
1. 10.21796 1
2. 2.39687 .866
3. 1.84975 .8165
4. 1.15361 .9221
5. .19046 .1627
6. .08276 .059
7. .06986 .0585
8. .0173 .0273
9. 8.43D-3 .0257
10. 6.66D-3 -2D-3 11.
2.76D-3 -.7772 12. 2.55D-3 -1.0375
13. 8.7D-4 -2.365 14.
5.6D-4 -.043 15.
0 -.733
16.-3.9D-4 -1.1081
The matrix is not positive definit. Cholesky decomposition
is not success-
Eigenvalues in per cent of trace = 16
1 .6386 2 .1498 3 .1156
4 .0721 5 .0119 6 5.2D-3
7 4.4D-3 8 1.1D-3 9 5D-4 10
4D-4 11 2D-4 12 2D-4
13 1D-4 14 0 15
0 16 0
[Intern: analysed: 10/22/02 23:21:46 PRG version
05/24/94 MA9.BAS
File = C:\OMI\NUMERIK\MATRIX\SMA\TRAPEZ32\TRAPEZ32.SMA
with data from C:\OMI\NUMERIK\MATRIX\SMA\TRAPEZ32\TRAPEZ32.D16
Date: 10/22/02 Time:23:21:46]
(5)
Values are computed and not measured with 2-digit-input-accuracy read
TRAPEZ32.D16 korrigiert with 2-digit-input
accuracy read
Samp Or MD NumS Condit Determinant
HaInRatio R_OutIn K_Norm C_Norm
32 16 0 --4 14892.5
0 7.01D-18 17952
0(8) -1(-1)
********** Summary of standard correlation matrix
analysis ***********
File = TRAPEZ32.D16 N-order= 16 N-sample= 32
Rank= 16 Missing data = 0
Positiv Definit=Cholesky successful________= No with 4 negat.
eigenvalue/s
HEVA: Highest eigenvalue abs.value_________=
10.215085044003685
LEVA: Lowest eigenvalue absolute value_____=
6.8592119996252823D-4
CON: Condition number HEVA/LEVA___________~=
14892.505209872116
DET: Determinant original matrix (OMIKRON)_=
9.0100922957000011D-22
DET: Determinant (CHOLESKY-Diagonal^2)_____= -999 (not
positive definit)
DET: Determinant (PESO-CHOLESKY)___________= -999 (not
positive definit)
DET: Determinant (product eigenvalues)_____=
9.0100922957003092D-22
DET: Determ.abs.val.(PESO prod.red.norms)__=
9.0100922957000008D-22
HAC: HADAMARD condition number_____________=
2.3482342345089274D-28
HCN: Heuristic condition |DET|CON__________=
6.0500850385650946D-26
D_I: Determinant Inverse absolute value____=
1.1098665442941606D+21
HDA: HADAMARD Inequality absolute value___<=
1.5824808367746779D+38
HIR: HADAMARD RATIO: D_I / HDA ____________=
7.01345961671313D-18
Highest inverse positive diagonal value____=
57.512212037
thus multiple r( 5.rest)_________________=
.991268071
and 1 multiple r > .99
Highest inverse negative diagonal value____= -8.589202343
thus multiple r( 1.rest)_________________=
1.056610262 (!)
and there are 12 multiple r > 1 (!)
Maximum range (upp-low) multip-r( 2.rest)_=
.171
LES: Numerical stability analysis:
Ratio maximum range output / input _______=
17951.973072121561
PESO-Analysis correlation least Ratio RN/ON=
4.51D-4 (<-> Angle = .03 )
Number of Ratios correlation RN/ON < .01__ =
8
PESO-Analysis Cholesky least Ratio RN/ON__ = (Not positiv definit)
Ncor L1-Norm L2-Norm Max
Min m|c| M|c| N_comp
s-S S-S
256 154.7 10.72
1 -.83 .578
.279 7140 .322 .231
class boundaries and distribution of the correlation coefficients
-1 -.8 -.6 -.4 -.2 0
.2 .4 .6 .8 1
2 10 10
22 12 16 24 32
46 82
i.Eigenvalue Cholesky i.Eigenvalue
Cholesky i.Eigenvalue Cholesky
1. 10.21509 1
2. 2.39196 .866
3. 1.85126 .8165
4. 1.15826 .92
5. .18978 .1697
6. .09108 .1099
7. .06916 .0399
8. .01877 -.0188
9. .01635 -.8712
10. .01019 -1.3064 11. 6.72D-3
-1.7335 12. 3.56D-3 -2.7824
13.-6.9D-4 -4.0284 14.-3.77D-3
-.1784 15.-6.87D-3 -1.515
16.-.01083 -1.7234
The matrix is not positive definit. Cholesky decomposition
is not success-
Eigenvalues in per cent of trace = 16
1 .6384 2 .1495 3 .1157
4 .0724 5 .0119 6 5.7D-3
7 4.3D-3 8 1.2D-3 9 1D-3 10
6D-4 11 4D-4 12 2D-4
13 0 14-2D-4 15-4D-4
16-7D-4
[Intern: analysed: 10/22/02 23:20:00 PRG version
05/24/94 MA9.BAS
File = C:\OMI\NUMERIK\MATRIX\SMA\TRAPEZ32\TRAPEZ32.SMA
with data from C:\OMI\NUMERIK\MATRIX\SMA\TRAPEZ32\TRAPEZ32.D16
Date: 10/22/02 Time:23:20:00]
(6)
Values measured with 2-digit input accuracy read
THURSTONE,L.L. (USA: The University of Chicago). "SECOND-ORDER FACTORS"
Psychometrika 9.2,1944, p. 96 Table 7. Bemerkung 1: die Tablele enthält
zwei Druckfehler (r14,16 und r15,16 sollten positiv sein).
6=12f) THPMF16.K16 raw scores measured with 2-digit input
accuray read
Samp Or MD NumS Condit Determinant
HaInRatio R_OutIn K_Norm C_Norm
32 16 0 --4 3711
-3.67 D-20 3.12 D-23 347573 0(8)
-1(-1)
Wir sehen hier einen
Widerspruch zwischen negativer Determinante und geradzahlig negativen Eigenwerte,
die zu einer positiven Determinante führen müßten. Daher
habe ich Determinante und Eigenwerte noch einmal mit dem genaueren Matlab
nachgerechnet, das für diese Matrix zu folgenden Ergebnissen
gelangt:
Det = -3.6706e-020, also gleicher Wert wie ihn das Omikron-Basisprogramm
berechnet. Zum Eigenwertvergleich siehe bitte unten
bei den Eigenwerten.
********** Summary of standard correlation matrix
analysis ***********
File = THPMF16.K16 N-order= 16 N-sample= 32
Rank= 16 Missing data = 0
Positiv Definit=Cholesky successful________= No with
4 negative eigenvalue/s
HEVA: Highest eigenvalue abs.value_________= 10.261
LEVA: Lowest eigenvalue absolute value_____= 0.002764723
CON: Condition number HEVA/LEVA___________~= 3711.28
DET: Determinant original matrix___________= D-20 -3.670
HAC: HADAMARD condition number_____________= D-27 8.551
HCN: Heuristic condition |DET|CON__________= D-24 9.890
D_I: Determinant Inverse absolute value____= 2.7243326243127375D+19
HDA: HADAMARD Inequation absolute value___<= 8.7270894616169875D+41
HIR: HADAMARD RATIO: D_I / HDA ____________= 3.1216966851259516D-23
Highest inverse positive diagonal value____= 637.130010907
thus multiple r( 6.rest)_________________= .999214923
and 10 multiple r > .99
Highest inverse negative diagonal value____= -9.745219835
thus multiple r( 12.rest)_________________= 1.05005448
(!)
and there are 2 multiple r > 1 (!)
Maximum range (upp-low) multip-r( 15.rest)_= .454
LES: Numerical stability analysis:
Maximum range input x(upper)-x(lower)____=
0.009
Maximum range output x(upper)-x(lower)____= 3128.15835
Ratio maximum range output / input _______= D +5
3.4
Mean absolute value of ranges output _____= 1118.18184
Ratio mean range output/ mean range input_= D +5
1.2
Sigma of mean (abs. value range output)___= 984.60139
Ncor L1-Norm L2-Norm Max
Min m|c| s|c|
Ncomp M-S S-S
256 156 10.79
1 -.84 .61
.537 120 .218 .244
class boundaries and distribution of the correlation-coefficients
-1 -.8 -.6 -.4 -.2 0
.2 .4 .6 .8 1
2 9 12
21 12 18 23 32
45 82
REMARK: The table contents two print errors (r14,16 and r15,16 have
to be positive)
Original input data with 2-digit-accuracy and read with 2-digit-accuracy
(for control here the analysed original matrix):
1 2 3
4 5 6 7
8 9 10 11 12
13 14 15 16
1 1 .5 .5 .32
.29 .58 .72 .49 .58 .45 .31 .66
.53 .76 -.35 .11
2 .5 1 .5 .32
.36 .42 .57 .49 .74 .67 .33 .54
.64 -.16 -.14 -.23
3 .5 .5 1 .32
.52 .42 .88 .82 .9 .45 .3
.78 .75 .19 -.84 -.72
4 .32 .32 .32 1 .95
.96 .65 .8 .61 .91 .98 .78
.82 .12 -.22 -.15
5 .29 .36 .52 .95 1
.9 .75 .9 .75 .9 .94
.84 .89 .05 -.37 -.31
6 .58 .42 .42 .96 .9
1 .78 .83 .7 .92 .94
.86 .86 .34 -.29 -.09
7 .72 .57 .88 .65 .75 .78
1 .95 .95 .74 .64 .95 .91
.39 -.69 -.46
8 .49 .49 .82 .8 .9
.83 .95 1 .93 .83 .78 .94
.95 .19 -.64 -.52
9 .58 .74 .9 .61 .75
.7 .95 .93 1 .79 .6
.9 .93 .11 -.64 -.57
10 .45 .67 .45 .91 .9 .92
.74 .83 .79 1 .9 .83
.9 .01 -.22 -.21
11 .31 .33 .3 .98 .94 .94
.64 .78 .6 .9 1 .77
.8 .11 -.12 -.09
12 .66 .54 .78 .78 .84 .86
.95 .94 .9 .83 .77 1
.97 .34 -.59 -.39
13 .53 .64 .75 .82 .89 .86
.91 .95 .93 .9 .8 .97 1
.12 -.52 -.44
14 .76 -.16 .19 .12 .05 .34 .39
.19 .11 .01 .11 .34 .12 1
-.28 -.34
15 -.35 -.14 -.84 -.22 -.37 -.29 -.69 -.64 -.64 -.22 -.12 -.59
-.52 -.28 1 -.76
16 .11 -.23 -.72 -.15 -.31 -.09 -.46 -.52 -.57 -.21 -.09 -.39 -.44
.34 .76 1
i.Eigenvalue Cholesky i.Eigenvalue
Cholesky i.Eigenvalue Cholesky
1. 10.26065 1
2. 2.17133 .866
3. 1.75624 .8165
4. 1.0651 .92
5. .88599 .1697
6. .07148 -5.6D-3
7. .0514 -.5981
8. .02466 -1.5864 9.
.014 -2.2646
10. 9.28D-3 -2.7013 11. 5.2D-3
-3.0387 12. 2.76D-3 -4.5821
13.-4.57D-3 -5.6998 14.-8.45D-3
-.4257 15.-.01274 -1.9947
16.-.22922 -1.8594
The matrix is not positive definit. Cholesky decomposition
is not successful (for detailed information Cholesky's diagonalvalues are
presented).
Anmerkung:
Bei den Eigenwerten kommt Matlab zu folgenden und vermutlich genaueren
Ergebnissen als das Omikron- Basic- Programm:
1. 10.2577
2. 2.1193
3. 1.5704
4. 0.8507 + 0.2465i 5.
0.8507 - 0.2465i 6. 0.2378
7. 0.0711
8. 0.0273
9. -0.0133
10. -0.0088
11. -0.0074
12. 0.0170
13. 0.0139
14. 0.0004
15. 0.0038
16. 0.0093
5.
Überblick der berechneten Trapezoid-Parameter (k)
Korrigierte Version
Thurstone's trapezoid raw scores with 3-digit-accuracy for view:
s1 s2 s3 s4 s5
s6 s7 s8
s9 s10 s11 s12
s13 s14 s15 s16
01 1 2 1 2 2
2.24 2.83 2.24 3.61 2.83
2.24 1 4 .5
2 1
02 1 2 1 4 4
4.12 4.47 4.12 5
4.47 4.12 2 8
.5 2 1
03 1 2 3 2 2.83 2.24
4.47 3.61 5.39 2.83 2.24
3 6 .5 .67
.33
04 1 2 3 4 4.47 4.12
5.66 5 6.4
4.47 4.12 6 12
.5 .67 .33
05 1 3 1 2 2
2.24 2.83 2.24 4.47 3.61
2.83 1 5 .33
3 1
06 1 3 1 4 4
4.12 4.47 4.12 5.66 5
4.47 2 10 .33 3
1
07 1 3 3 2 2.83 2.24
4.47 3.61 6.32 3.61 2
3 7 .33 1
.33
08 1 3 3 4 4.47 4.12
5.66 5 7.21 5
4 6 14
.33 1 .33
09 2 2 1 2 2.24 2.83
3.61 2.24 3.61 2.83 2.24
1 5 1 2
2
10 2 2 1 4 4.12 4.47
5 4.12 5
4.47 4.12 2 10
1 2 2
11 2 2 3 2 2.24 2.83
5.39 3.61 5.39 2.83 2.24
3 7 1 .67
.67
12 2 2 3 4 4.12 4.47
6.4 5 6.4
4.47 4.12 6 14
1 .67 .67
13 2 3 1 2 2.24 2.83
3.61 2.24 4.47 3.61 2.83
1 6 .67 3
2
14 2 3 1 4 4.12 4.47
5 4.12 5.66 5
4.47 2 12 .67 3
2
15 2 3 3 2 2.24 2.83
5.39 3.61 6.32 3.61 2
3 8 .67 1
.67
16 2 3 3 4 4.12 4.47
6.4 5 7.21
5 4 6
16 .67 1 .67
17 2 3 3 3 3.16 3.61
5.83 4.24 6.71 4.24 3
4.5 12 .67 1 .67
18 2 3 3 5 5.1
5.39 7.07 5.83 7.81 5.83
5 7.5 20 .67
1 .67
19 2 3 5 3 4.24 3.61
7.62 5.83 8.54 4.24 3.61
7.5 15 .67 .6 .4
20 2 3 5 5 5.83 5.39
8.6 7.07 9.43 5.83
5.39 12.5 25 .67 .6 .4
21 2 4 3 3 3.16 3.61
5.83 4.24 7.62 5
3.16 4.5 13.5 .5 1.33 .67
22 2 4 3 5 5.1
5.39 7.07 5.83 8.6
6.4 5.1 7.5 22.5 .5
1.33 .67
23 2 4 5 3 4.24 3.61
7.62 5.83 9.49 5
3.16 7.5 16.5 .5 .8 .4
24 2 4 5 5 5.83 5.39
8.6 7.07 10.3 6.4
5.1 12.5 27.5 .5 .8 .4
25 3 3 3 3 3
4.24 6.71 4.24 6.71 4.24
3 4.5 13.5 1
1 1
26 3 3 3 5 5
5.83 7.81 5.83 7.81 5.83
5 7.5 22.5 1
1 1
27 3 3 5 3 3.61 4.24
8.54 5.83 8.54 4.24 3.61
7.5 16.5 1 .6 .6
28 3 3 5 5 5.39 5.83
9.43 7.07 9.43 5.83 5.39
12.5 27.5 1 .6 .6
29 3 4 3 3 3
4.24 6.71 4.24 7.62 5
3.16 4.5 15 .75 1.33 1
30 3 4 3 5 5
5.83 7.81 5.83 8.6
6.4 5.1 7.5 25
.75 1.33 1
31 3 4 5 3 3.61 4.24
8.54 5.83 9.49 5
3.16 7.5 18 .75 .8
.6
32 3 4 5 5 5.39 5.83
9.43 7.07 10.3 6.4
5.1 12.5 30 .75 .8
.6
6.
OMIKRON-Basic-Programm zur Berechnung der Parameter des THURSTONEschen
Trapezoids (k)
REM TRAPEZ.BAS (THURSTONE's trapezoid)
' 20.10.2002 R.Sponsel D-91052 Erlangen, A12 und
A13
korrigiert
COMPILER "TRACE ON"
DEFDBL "X"
S=16:Z=32: DIM Xd#(S,Z)
OPEN "I",1,"C:\OMI\NUMERIK\MATRIX\URDAT\THURS\TH429_4.N32"
FOR I=1 TO 4'
trapezoid parameter a,b,c,h read
FOR J=1 TO Z
INPUT #1,Xd#(I,J)
NEXT J
NEXT I: CLOSE 1
OPEN "O",1,"C:\OMI\NUMERIK\MATRIX\URDAT\THURS\TH429_32.N16"
OPEN "O",2,"C:\OMI\NUMERIK\MATRIX\URDAT\THURS\TH429_32.TAB"
FOR I=5 TO S
FOR J=1 TO Z
A#=Xd#(1,J):B#=Xd#(2,J):C#=Xd#(3,J):H#=Xd#(4,J)
IF I=5 THEN Xd#(I,J)= SQR((C#-A#)^2+H#^2)
IF I=6 THEN Xd#(I,J)= SQR(A#^2+H#^2)
IF I=7 THEN Xd#(I,J)= SQR((A#+C#)^2+H#^2)
IF I=8 THEN Xd#(I,J)= SQR(C#^2+H#^2)
IF I=9 THEN Xd#(I,J)= SQR((C#+B#)^2+H#^2)
IF I=10 THEN Xd#(I,J)= SQR(B#^2+H#^2)
IF I=11 THEN Xd#(I,J)= SQR((C#-B#)^2+H#^2)
IF I=12 THEN Xd#(I,J)=(((2*C#+A#+B#)/2)*H#)-((A#+B#+C#)/2)*H#
IF I=13 THEN Xd#(I,J)=((A#*B#+C#)/2)*H#
IF I=14 THEN Xd#(I,J)=A#/B#
IF I=15 THEN Xd#(I,J)=B#/C#
IF I=16 THEN Xd#(I,J)=A#/C#
NEXT J
NEXT I
FOR I=1 TO S
FOR J=1 TO Z
PRINT #1,Xd#(I,J)
NEXT J
NEXT I: CLOSE 1
PRINT #2,"Thurstone's trapezoid raw scores with 3-digit-accuracy
for view:"
FOR I=1 TO Z
FOR J=1 TO S
PRINT #2, TAB (7*(J-1)); INT(Xd#(J,I)*10^2+.5)/10^2;
NEXT J: PRINT #2
NEXT I
CLOSE 2
END
Fehlerhinweis:
Ich verdanke den Fehlerhinweis bei der Thurstone'schen Trapezoidberechnung
für die mißverständlich ausgewiesene Area
12 und Area 13 Hermann Kremer und Gottfried Helms
aus der Newsgroup de.sci.mathematik, in der ich nachfragte wie der
fünfte, 'zahlengefühlsmäßig' relativ große Eigenwert
der ersten gerechneten Version mit 17stelliger Genauigkeit von 0,370
erklärt werden könnte. Nach Beseitigung des Fehlers fiel dieser
Eigenwert auf relativ unauffällige 0,190
. Ich hoffe ich erliege hier keinem psychologischen ex post facto Phänomen
;-). Der relativ große Eigenwert fiel mir damals nicht auf, weil
diese Studie das Hauptinteresse verfolgte, die Indefinitheit der Matrix,
also die Produktion von negativen Eigenwerten, als Folge der Kombination
von Kollinearität und Rundungsfehlern durch Modellbildung zu beweisen.
Der relativ große Eigenwert fiel mir erst auf, als ich jüngst
die Reproduktionsmatrix aus den Faktoren
Thurstone's rückrechnete. Die Fehlerbearbeitung motivierte mich, zu
vergleichen, wie dieser Fehler sich auf die Qualität der Reproduktionsgüte
zwischen "gemessen" versus berechnet der rückgerechneten Matrizen
auswirkt. Das ist hier dokumentiert.
Interessant ist jedenfalls, daß die Meßfehler hier nur geringe
Auswirkungen auf die Eigenwerte haben, was gegen Screetest
und Kaisers grobschlächtiges
Kriterium spricht. Der Fehler hat glücklicherweise keine Auswirkungen
auf die wichtigen Aussagen, Schlußfolgerungen und Interpretation,
weil auch die fehlerhafte Berechnung die entsprechenden Parameter im Thurstone'schen
Sinne berücksichtigt, obwohl die Auswirkungen auf die Korrelationskoeffizienten
(Zeile/Spalte 12,13) teilweise erheblich sind (Residuen = Fehlerbehaftete
Korrelationsmatrix - Richtige Korrelationsmatrix):
Residual-Analysis:
Mean= .04556031 Sigma= .11576164 Maximum
range= .50578127 (r4.13)
Matrix residuals (whole matrix inclusive diagonal):
Mean absolute values of residuals = .045560306322708245
Sigma absolute values of residuals = .11576164489090045
Maximum range absolute values = .50578127183620955
(r4.13)
Matrix residuals upper triangular matrix without diagonal:
Mean absolute values of residuals = .048597660077555462
Sigma absolute values of residuals = .1189392
Maximum range absolute values = .50578127183620955
(r4.13)
Matrix of residuals
0 0 0
0 0 0
0 0 0
0 0 -.09 .103
0 0 0
0 0 0
0 0 0
0 0 .001 .014
0 0 0
0 0 0
0 0 0
0 0 -.085 -.243
0 0 0
0 0 0
0 0 0
0 0 .452 .506
0 0 0
0 0 0
0 0 0
0 0 .366 .337
0 0 0
0 0 0
0 0 0
0 0 .356 .461
0 0 0
0 0 0
0 0 0
0 0 .063 .046
0 0 0
0 0 0
0 0 0
0 0 .185 .122
0 0 0
0 0 0
0 0 0
0 0 .070 -.007
0 0 0
0 0 0
0 0 0
0 0 .352 .400
0 0 0
0 0 0
0 0 0
0 0 .424 .477
-.09 .001 -.085 .452 .366 .356
.063 .185 .070 .352 .424 0
-.041
.103 .014 -.243 .506 .337 .461
.046 .122 -.007 .400 .477 -.041 0
0 0 0
0 0 0
0 0 0
0 0 -.1 .102
0 0 0
0 0 0
0 0 0
0 0 .012 .158
0 0 0
0 0 0
0 0 0
0 0 -.038 .22
Ausblick: Inzwischen bin ich mit dem Thurstone'schen Trapezoiden so
vertraut, daß ich mit ihm eine realistische Fehlersimulationsstudie
(N=300) plane, um empirische Information darüber zu erhalten, wie
verändert sich die Faktorenstruktur und die Eigenwerte des Trapezoiden,
wenn man mit diesem oder jenem Fehlermodell arbeitet.
Querverweise:
Erste, fehlerhafte Trapezoid-Studie (K7_3.htm)
Thurstone Biographie
Aus 4-Faktoren rückgerechnete
Thurstone'schen Trapezoid Korrelationsmatrix*
Standard-Matrix-Analyse der Primary
Mental Abilities von Thurstone, L. L. (1938).
Kritik der Handhabung
der Faktorenanalyse
Für NichtmethodikerInnen: worauf kommt
es an bei Korrelationsmatrizen
Für professionell Interessierte: Abkürzungen,
Definition, Erklärung und Bedeutung zur
Standard- (Korrelations)- Matrix- Analyse (SMA)
Gesamtzusammenfassung:
"Numerisch instabile Matrizen und Kollinearität in der Psychologie"
Hintergrund
und Entstehungsgeschichte der Arbeit "Numerisch instabile Matrizen und
Kollinearität in der Psychologie"
Wird im Laufe der Zeit fortgesetzt, ergänzt und erweitert
FN01
Sponsel, Rudolf & Hain, Bernhard (1994). Numerisch instabile Matrizen
und Kollinearität in der Psychologie. Diagnose, Relevanz & Utilität,
Frequenz, Ätiologie, Therapie. Ill-Conditioned Matrices and
Collinearity in Psychology. Deutsch-Englisch. Übersetzt von Agnes
Mehl. Kapitel 6 von Dr. Bernhard Hain: Bemerkungen über Korrelationsmatrizen.
Erlangen: IEC-Verlag [ISSN-0944-5072 ISBN 3-923389-03-5]. Aktueller
Preis: https://ww.iec-verlag.de
Zitierung
Sponsel, Rudolf (DAS).
Korrigierte Dokumentation
Rundungsfehler & Kollinearität
am THURSTONEschen Trapezoid- Beispiel. Abteilung: Numerisch instabile Matrizen
und Kollinearität in der Psychologie - Ill-Conditioned Matrices and
Collinearity in Psychology - Diagnose, Relevanz & Utilität,
Frequenz, Ätiologie, Therapie. IP-GIPT.
Erlangen: https://www.sgipt.org/wisms/nis/k7/K7_3k.htm
Copyright & Nutzungsrechte
Diese Seite darf von jeder/m in nicht-kommerziellen
Verwertungen frei aber nur original bearbeitet und nicht inhaltlich
verändert und nur bei vollständiger Angabe der Zitierungs-Quelle
benutzt werden. In Streitfällen gilt der Gerichtsstand Erlangen als
akzeptiert.
Ende_
Trapezoid
(7.3korrigiert)
_Überblick
_ Relativ
Aktuelles _ Rel.
Beständiges _ Titelblatt
_ Konzept
_ Archiv
_ Region
_ Service
iec-verlag _ Mail:
_ Sekretariat@sgipt.org
_ _
__
_Wichtige
Hinweise zu Links und Empfehlungen