IP-GIPT DAS=06.12.2005 Internet-Erstausgabe, letzte Änderung TMJ
Impressum: Diplom-PsychologInnen Irmgard Rathsmann-Sponsel und Dr. phil. Rudolf Sponsel
Stubenlohstr. 20 D-91052 Erlangen Mail:_sekretariat@sgipt.org_
Anfang_ Coombs 1941 _ Überblick _Relativ Aktuelles _Rel. Beständiges _Titelblatt _ Konzept _ Archiv _ Region _ Service iec-verlag _Mail: Sekretariat@sgipt.org _ Zitierung & Copyright _ _ Wichtige Hinweise zu Links und Empfehlungen
Willkommen in der Abteilung Wissenschaftstheorie, Methodologie und Statistisch-Mathematische Methoden in der Allgemeinen und Integrativen Psychologie, Psychodiagnostik und Psychotherapie hier zum Thema Numerisch instabile Matrizen imd Kollinearität in der Psychologie und Psychotherapie:
Standard-Matrix-Analyse
Coombs, C.H. (1941). A criterion for significant common factor variance. Psychometrika 6, 267-272.
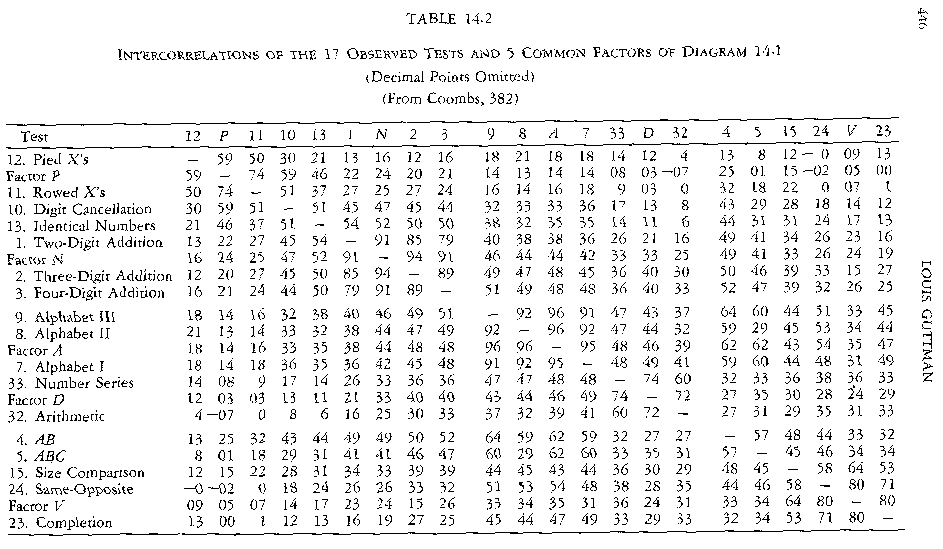
In: Guttman, Louis (1966). Order Analysis Of Correlation Matrices, p. 446.
In: Cattell, Raymond B. (1966, Ed.). Handbook Of Multivariate Experimental Psychology. Chicago: Rand & McNally.
| Zusammenfassung - Summary - Abstract: Die Matrix vom Coombs
ist nicht positiv semidefinit, sie ist indefinit mit zwei größeren
negativen negativen Eigenwerten (-.02013 und -.07025) entgleist,
die durch Rundungsfehler nicht erklärbar sind. Das heißt, die
Korrelationen müssen in unzulässiger Weise gewonnen worden
sein. Damit ist die phänomenologisch erscheinende Matrix im
mathematischen Sinne keine "genotypische" (echte) Korrelationsmatrix
(Produkt-Moment oder Bravais Pearson). Die Entgleisung zeigt sich sehr
drastisch durch einen multiplen Korrelationskoeffizienten von 1.5715
(!) und insgesamt 5
multiplen Korrelationskoeffizienten > 1.
Samp _Ord_ MD_ NumS_ Condition_Determinant_HaInRatioR_ OutInK_ Norm_ C Norm ? 22 -1 --2 452.8 0 8.34D-11 44.8 .013(0) -1(-1) |
|
|
********** Summary
of standard correlation matrix analysis ***********
File = coombs41.k22 N-order= 22 N-sample=?
Rank= 22 Missing data = ?
Positiv Definit=Cholesky successful________= No with 2 negat.
eigenvalue/s
HEVA: Highest eigenvalue abs.value_________=
9.1144109647408429
LEVA: Lowest eigenvalue absolute value_____=
.020127036090946055
CON: Condition number HEVA/LEVA___________~=
452.84417057516329
DET: Determinant original matrix (OMIKRON)_=
6.0403604739505258D-11
DET: Determinant (CHOLESKY-Diagonal^2)_____= -999 (not
positive definit)
DET: Determinant (PESO-CHOLESKY)___________= -999 (not
positive definit)
DET: Determinant (product eigenvalues)_____=
6.0403604739505181D-11
DET: Determ.abs.val.(PESO prod.red.norms)__=
6.0403604739505258D-11
HAC: HADAMARD condition number_____________=
4.1650445190459257D-18
HCN: Heuristic condition |DET|CON__________=
1.3338717524570506D-13
D_I: Determinant Inverse absolute value____=
16555303352
HDA: HADAMARD Inequality absolute value___<=
1.9843086250570484D+20
HIR: HADAMARD RATIO: D_I / HDA ____________=
8.3431091024188185D-11
Highest inverse positive diagonal value____=
14.081193508
thus multiple r( 10.rest)_________________=
.963837794
Highest inverse negative diagonal value____= -.680449043
thus multiple r( 22.rest)_________________=
1.571501754 (!)
and there are 5 multiple r > 1 (!)
Maximum range (upp-low) multip-r( 22.rest)_=
2.509
LES: Numerical stability analysis:
Ratio maximum range output / input _______=
44.751549794654189
PESO-Analysis correlation least Ratio RN/ON=
.013422 (<-> Angle = .77 )
Number of Ratios correlation RN/ON < .01__ =
0
PESO-Analysis Cholesky least Ratio RN/ON__ = (Not positiv definit)
Ncor
L1-Norm L2-Norm Max Min m|c|
s|c| N_comp M-S S-S
484 190.2
10.17 1 -.07
.364 .209 26565
.231 .186
class boundaries and distribution of the correlation coefficients
-1 -.8 -.6 -.4 -.2 0
.2 .4 .6 .8 1
0 0 0
0 12 98 170 138 22
44
Original data with 2, input read with 2, computet with
19,
and showed with 2 digit accuracy (for control here
the analysed original matrix):
1 .59 .5 .3
.21 .13 .16 .12 .16 .18 .21 .18
.18 .14 .12 .04
.59 1 .74 .59 .46
.22 .24 .2 .21 .14 .13 .14
.14 .08 .03 -.07
.5 .74 1 .51 .37
.27 .25 .27 .24 .16 .14 .16 .18
.09 .03 0
.3 .59 .51 1 .51
.45 .47 .45 .44 .32 .33 .33 .36
.17 .13 .08
.21 .46 .37 .51 1
.54 .52 .5 .5 .38 .32 .35
.35 .14 .11 .06
.13 .22 .27 .45 .54 1
.91 .85 .79 .4 .38 .38 .36
.26 .21 .16
.16 .24 .25 .47 .52 .91
1 .94 .91 .46 .44 .44 .42
.33 .33 .25
.12 .2 .27 .45 .5
.85 .94 1 .89 .49 .47 .48
.45 .36 .4 .3
.16 .21 .24 .44 .5 .79
.91 .89 1 .51 .49 .48 .48
.36 .4 .33
.18 .14 .16 .32 .38 .4
.46 .49 .51 1 .92 .96 .91
.47 .43 .37
.21 .13 .14 .33 .32 .38
.44 .47 .49 .92 1 .96 .92
.47 .44 .32
.18 .14 .16 .33 .35 .38
.44 .48 .48 .96 .96 1 .95
.48 .46 .39
.18 .14 .18 .36 .35 .36
.42 .45 .48 .91 .92 .95 1
.48 .49 .41
.14 .08 .09 .17 .14 .26
.33 .36 .36 .47 .47 .48 .48 1
.74 .6
.12 .03 .03 .13 .11 .21
.33 .4 .4 .43 .44 .46 .49
.74 1 .72
.04 -.07 0 .08 .06 .16
.25 .3 .33 .37 .32 .39 .41
.6 .72 1
.13 .25 .32 .43 .44 .49
.49 .5 .52 .64 .59 .62 .59
.32 .27 .27
.08 .01 .18 .29 .31 .41
.41 .46 .47 .6 .29 .62 .6
.33 .35 .31
.12 .15 .22 .28 .31 .34
.33 .39 .39 .44 .45 .43 .44 .36
.3 .29
0 -.02 0 .18 .24
.26 .26 .33 .32 .51 .53 .54 .48
.38 .28 .35
.09 .05 .07 .14 .17 .23
.24 .15 .26 .33 .34 .35 .31 .36
.24 .31
.13 0 .01 .12 .13
.16 .19 .27 .25 .45 .44 .47 .49
.33 .29 .33
.13 .08 .12 0 .09
.13
.25 .01 .15 -.02 .05 0
.32 .18 .22 0 .07
.01
.43 .29 .28 .18 .14 .12
.44 .31 .31 .24 .17 .13
.49 .41 .34 .26 .23 .16
.49 .41 .33 .26 .24 .19
.5 .46 .39 .33 .15 .27
.52 .47 .39 .32 .26 .25
.64 .6 .44 .51 .33 .45
.59 .29 .45 .53 .34 .44
.62 .62 .43 .54 .35 .47
.59 .6 .44 .48 .31 .49
.32 .33 .36 .38 .36 .33
.27 .35 .3 .28 .24 .29
.27 .31 .29 .35 .31 .33
1 .57 .48 .44 .33
.32
.57 1 .45 .46 .34
.34
.48 .45 1 .58 .64
.53
.44 .46 .58 1 .8
.71
.33 .34 .64 .8 1
.8
.32 .34 .53 .71 .8 1
i.Eigenvalue Cholesky i.Eigenvalue
Cholesky i.Eigenvalue Cholesky
1. 9.11441 1
2. 3.05858 .8074
3. 1.95686 .668
4. 1.65374 .7967
5. 1.46789 .8355
6. .81712 .7963
7. .65813 .4056
8. .51057 .3266
9. .48039 .3872
10. .41222 .8242
11. .3872 .3799
12. .36244 .1931
13. .27688 .2961
14. .2338 .8485
15. .20248 .5943
16. .17469 .6333
17. .16373 .6816
18. .07963 -1.0387
19. .05279 .6538
20. .02683 .5897
21.-.02013 -.0641
22.-.07025 -.2761
The matrix is not positive definit. Cholesky decomposition
is not successful.
Eigenvalues in per cent of trace = 21.999999999999999
1 .4143 2 .139 3 .0889
4 .0752 5 .0667 6 .0371
7 .0299 8 .0232 9 .0218
10 .0187 11 .0176 12 .0165
13 .0126 14 .0106 15 9.2D-3 16 7.9D-3 17 7.4D-3
18 3.6D-3
19 2.4D-3 20 1.2D-3 21-9D-4 22-3.2D-3
analysed: 12/05/05 22:04:36 PRG version 05/24/94
MA9.BAS
[Interne Verwaltung: File = C:\OMI\NUMERIK\MATRIX\SMA\coombs41\coombs41.SMA
with data from C:\OMI\NUMERIK\MATRIX\SMA\coombs41\coombs41.k22
Date: 12/05/05 Time:22:04:36]
Multiple correlations of original matrix
r1.rest = .60830421637969243
r2.rest = .83150244686471082
r3.rest = .72255511914761746
r4.rest = .72499976754739536
r5.rest = .69677119296304202
r6.rest = .92744703638335478
r7.rest = imaginary with radicand -1.1840834886631961
r8.rest = 1.0333091726902099
r9.rest = .92863322400750881
r10.rest = .96383779368842841
r11.rest = 1.0757284750945087
r12.rest = .96331337753423112
r13.rest = .9413174325563095
r14.rest = .78431383745618764
r15.rest = .83369212519929888
r16.rest = .73175565639119942
r17.rest = .74144383636336374
r18.rest = 1.4981160629242179
r19.rest = .45026634182605156
r20.rest = .707070626062042
r21.rest = 1.0673517927832424
r22.rest = 1.5715017536363515
GIF-Faksimile aus Guttman in Cattell (1966)
___
FN01 Sponsel, Rudolf & Hain, Bernhard (1994). Numerisch instabile Matrizen und Kollinearität in der Psychologie. Diagnose, Relevanz & Utilität, Frequenz, Ätiologie, Therapie. Ill-Conditioned Matrices and Collinearity in Psychology. Deutsch-Englisch. Übersetzt von Agnes Mehl. Kapitel 6 von Dr. Bernhard Hain: Bemerkungen über Korrelationsmatrizen. Erlangen: IEC-Verlag [ISSN-0944-5072 ISBN 3-923389-03-5]. Aktueller Preis: www.iec-verlag.de.
Bd. 2.: Sponsel, R. (2005). Fast- Kollinearität in Korrelationsmatrizen mit Eigenwert-Analysen erkennen Ergänzungsband - Band II zu
Numerisch instabile Matrizen und Kollinearität in der Psychologie.
Änderungen wird gelegentlich überarbeitet, ergänzt und vertieft * Anregungen und Kritik willkommen
00.00.00
Standort: Vorlage SMA
Überblicks- und Verteilerseite: Numerisch instabile Matrizen und Kollinearität in der Psychologie - Ill-Conditioned Matrices and Collinearity in Psychology - Diagnose, Relevanz & Utilität, Frequenz, Ätiologie, Therapie.
* Überblick Statistik in der IP-GIPT * Überblick Wissenschaft in der IP-GIPT. * Überblick Faktorenananlyse *
*
| Suchen in der IP-GIPT,
z.B. mit Hilfe von "google": <suchbegriff>
site:www.sgipt.org
z.B. Numerisch instabile Matrizen site:www.sgipt.org. * Korrelation site:www.sgipt.org |
Dienstleistungs-Info.
*
Zitierung
Sponsel, Rudolf (DAS). Eine indefinite Korrelationsmatrix von Coombs, C.H. (1941). A criterion for significant common factor variance. Psychometrika 6, 267-272. Aus der Abteilung Numerisch instabile Matrizen und Kollinearität in der Psychologie - Ill-Conditioned Matrices and Collinearity in Psychology - Diagnose, Relevanz & Utilität, Frequenz, Ätiologie, Therapie. IP-GIPT. Erlangen: https://www.sgipt.org/wisms/nis/sma/coombs41.htm
Copyright & Nutzungsrechte
Diese Seite darf von jeder/m in nicht-kommerziellen Verwertungen frei aber nur original bearbeitet und nicht inhaltlich verändert und nur bei vollständiger Angabe der Zitierungs-Quelle benutzt werden. Das Einbinden in fremde Seiten oder Rahmen, die die Urheberschaft der IP-GIPT nicht jederzeit klar erkennen lassen, ist nicht gestattet. Sofern die Rechte anderer berührt sind, sind diese dort zu erkunden. Sollten wir die Rechte anderer unberechtigt genutzt haben, bitten wir um Mitteilung. Soweit es um (längere) Zitate aus ... geht, sind die Rechte bei/m ... zu erkunden oder eine Erlaubnis einzuholen.
Ende_ Coombs 1941 _Überblick _Relativ Aktuelles _Rel. Beständiges _Titelblatt _ Konzept _Archiv _Region _Service iec-verlag _Mail: Sekretariat@sgipt.org _ _ Wichtige Hinweise zu Links und Empfehlungen